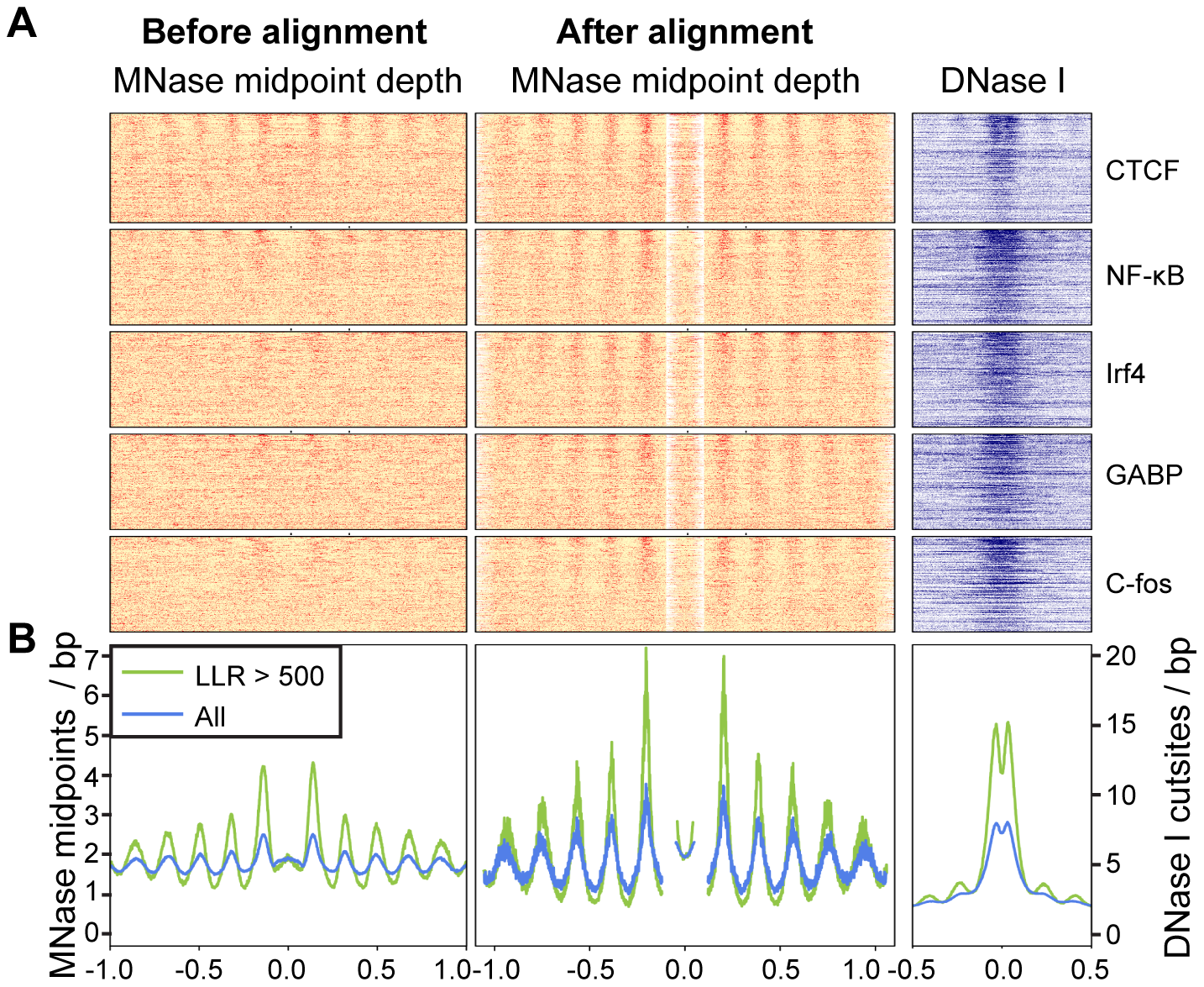
library(zoo) # install package 'zoo' using install.packages("zoo")
color <- colorRampPalette(c("white", "red"), space = "rgb")(100)
x <- rollapply(data, width=20, mean, by=20, by.row=TRUE)
y <- rollapply(data_shifted, width=20, mean, by=20, by.row=TRUE)
layout(matrix(c(1,2,3,4), nrow=2, ncol=2), heights=c(1.5,1.5))
par(mar=c(0,5,0,0.5), oma=c(5,0,2,0))
image(t(x), col=color, xaxt="n", yaxt="n", bty="n")
plot(seq(-990, 990, 10), colMeans(data), type="l", lwd=2, ylab="", xlab="", bty="n", ylim=c(0,0.4))
image(t(y), col=color, xaxt="n", yaxt="n", bty="n")
plot(seq(-940, 940, 10), colMeans(data_shifted), type="l", lwd=2, ylab="", xlab="", bty="n",col="blue", ylim=c(0,0.4))
par(new=T)
plot(seq(-940, 940, 10), colSums(data_shifted*P_shifted)/sum(P_shifted), type="l", lwd=2, ylab="", xlab="", bty="n",col="green", ylim=c(0,0.4))
legend("topleft", legend=c("class 1","ALL"), lty=1, col=c("green", "blue"), bty="n", lwd=c(5,5))