Lausanne, 27 April - 1 May 2015

Have a look at the Figure legend and the Methods section of the corresponding paper. However, you are not asked to precisely follow the data analysis protocol of the others. Just use the relevant sequence conservation and SNP tracks from the ChIP-Seq server to produce figures of the same kind. Once you have the results, you may ask yourself whether you agree with the interpretation by the authors:
CHR START END STRAND SEQ TF PWM_SCORE ... 2L 18564 18574 1 TAGTGTGCCCG trx_disc1 6.06 ... 2L 18917 18927 1 CAGTGTGACCA trx_disc1 8.2 ... 2L 21369 21379 1 TAGTGTGCCCG trx_disc1 6.06 ... 2L 21742 21752 1 CAGTGTGACCA trx_disc1 8.2 ... 2L 22068 22078 1 CAGTGTGGACG trx_disc1 6.15 ... 2L 28228 28237 -1 AGCTACCTGT hb_disc1 6.16 ...The genomic coordinates refer to the D. melanogaster genome assembly dm3. The file contains binding sites for 10 different transcription factors identified by the codes: bin_ef, cnc_disc1, h_known1, hb_disc1, hkb_known2, mod_disc3, tin_ef, trx_disc1 and twi_ef- We are only interested in twi_ef (Twist), bin_ef (Binou) and tin_ef (Tinman).
Several editing operations need be applied to transform the lines of this table into a valid BED file that can be uploaded to the ChIP-seq server:
bed = read.table("spivakov12_S3.txt", as.is=1, header=T)
bed[,1]=paste("chr", as.character(bed[,1]), sep="")
bed[,2]=bed[,2]-1
bed[which(bed[,4] == 1),4] = "+"
bed[which(bed[,4] == -1),4] = "-"
write.table(bed[which(bed[,6] == "twi_ef"),c(1,2,3,5,6,4)],
"twi_ef.bed", quote=F, sep="\t", row.names=F, col.names=F, eol="\n")
write.table(bed[which(bed[,6] == "tin_ef"),c(1,2,3,5,6,4)],
"tin_ef.bed", quote=F, sep="\t", row.names=F, col.names=F, eol="\n")
write.table(bed[which(bed[,6] == "bin_ef"),c(1,2,3,5,6,4)],
"bin_ef.bed", quote=F, sep="\t", row.names=F, col.names=F, eol="\n")
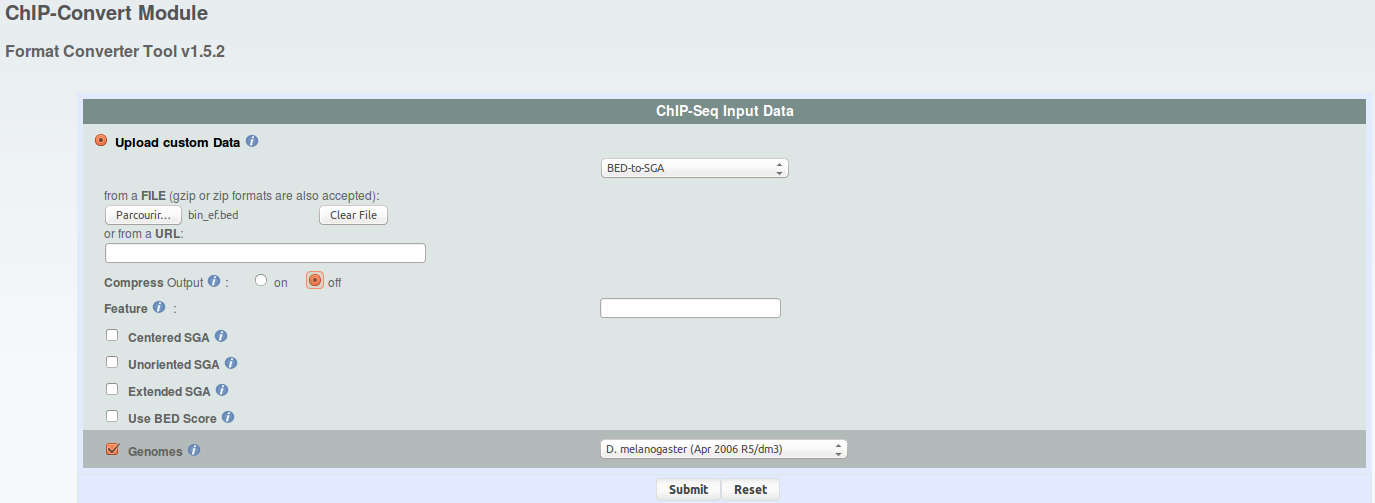
The bed files produced in this way can be uploaded to
ChIP-Convert. Set
the parameters as shown on the image.
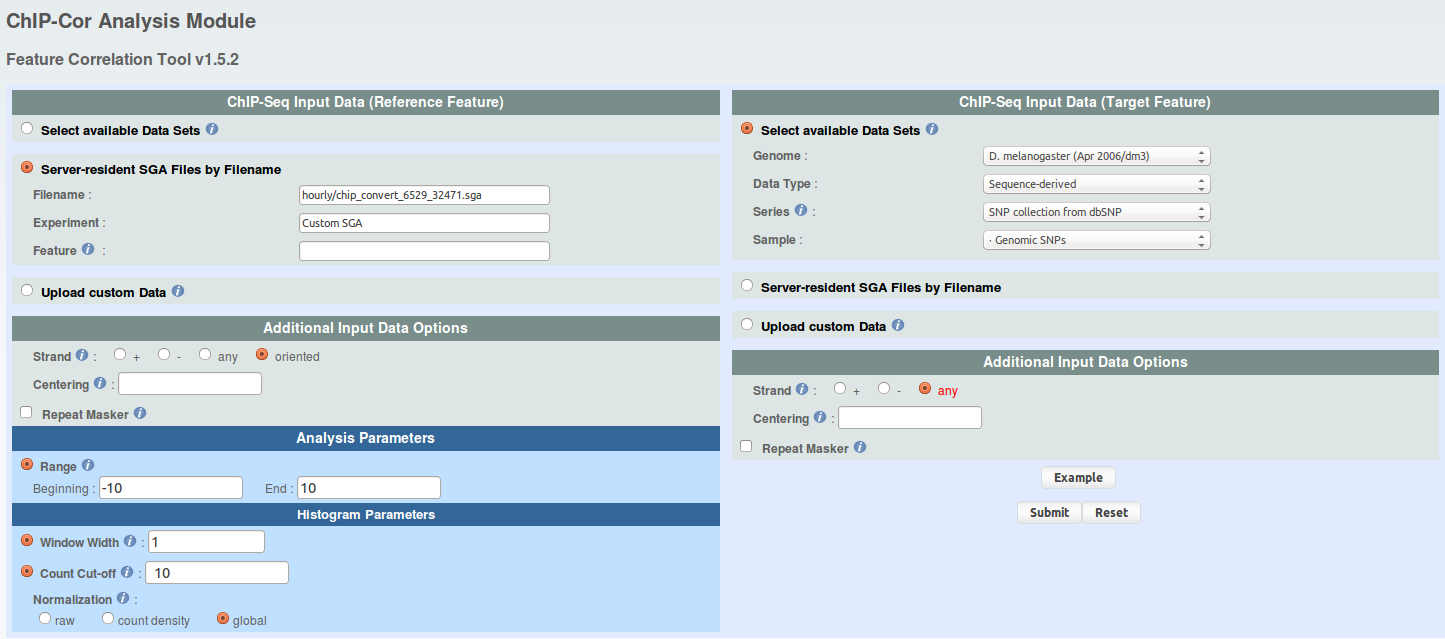
From the results page, you can directly navigate to the ChIP-Cor server and set the parameters as on the image below. Since the binding motifs are asymmetric and the binding sites may occur in either orientation, you should specify "strand oriented" for the reference feature. The count cut-off may invariantly be set to 10 since as all relevant tracks have count values of at most 10. The relevant target feature tracks for this exercise are: