Introduction
The motif databases used by our web resources have been downloaded from The MEME Suite website (latest MEME motif databases update of 07 Dec 2017).
Matrices are provided in two formats:
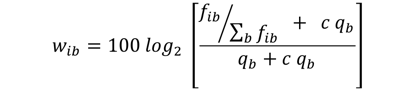
where fib is the frequency of base b at PWM position i, qb is the background frequency of base b, and c is the fraction of pseudo-counts added to the observed base frequencies. Unless specified otherwise, background letter frequencies are those from uniform background (A 0.25000 C 0.25000 G 0.25000 T 0.25000). Weights are rounded to nearest integers to allow for efficient computation of the probability distribution for scores expected from random sequences. For more details, visit the MEME website.
In most cases, the MEME motif database file provides letter-probability matrices. Alternatively, both letter-probability and log-odds matrices are included in the original MEME file. The letter-probability matrix is a table of probabilities where the rows are positions in the motif and the columns are letters in the alphabet that are ordered alphabetically. Therefore, for DNA motifs the colums are in the order A, C, G, T. Each row reports the probability of each letter in the alphabet (the sum of all probabilities in the row must be 1). Here below is an example of MEME-formatted DNA motif's letter-probability matrix:
We extract the probability matrices from the original MEME file, modify the header and generate the corresponding log-odds matrix using the background frequencies provided by MEME and a pseudo-count fraction of zero. As log of zero is not defined, we give it a value of -10000. Considering the previous example, the corresponding local formats for letter-probability and log-odds are shown here:
The new matrix header results from merging the original MEME header into a single line that includes the unique MEME motif identifier and name. For matrix conversion we use ad hoc scripts that implement the same mathematics as the one used by the MEME software. In the case that MEME database files provide both letter-probability and log-odds matrices, we just extract the matrix lines and modify the header according to the local format convention.
|
PWM Collections
Text taken from JASPAR DOWNLOAD, and partially modified. The JASPAR CORE database contains a curated, non-redundant set of 746 profiles derived from published collections of experimentally defined transcription factor (TF) binding sites for multi-cellular eukaryotes. The TF binding profiles were historically determined from SELEX experiments. More recent profiles are derived from high-throughput techniques such as ChIP-sequencing, Protein Binding Microarray, or High-Throughput SELEX. The prime difference to similar resources (TRANSFAC, etc) consist of the open data acess, non-redundancy and quality. Motif length is between 6 and 21 bp (average width 11.8). References:
Text taken from JASPAR Clusters, and converted to MEME-like format. This collection includes 79 root motifs obtained by clustering the 579 profiles from the JASPAR CORE (2018) vertebrates source database, using the RSTA matrix-clustering tool. matrix-clustering is a program that clusters similar transcription factor binding motifs (TFBMs) into multiple trees, and automatically creates non-redundant TFBM collections. A key feature of matrix-clustering is its dynamic visualisation of aligned TFBMs, and its capability to simultaneously treat multiple collections from various sources. Motif length is between 7 and 25 bp (average width 15.7). References:
Text taken from MEME, and partially modified. The Jolma 2013 Human and Mouse database contains a curated set of non-redundant motifs describing 235 distinctly different binding specificities. The sequence-specific binding of human TFs has been studied using high-throughput SELEX and ChIP sequencing. Motif width is between 7 and 23 (average width 12.7). References:
Text taken from MEME, and partially modified. The Jolma 2013 Human and Mouse database contains a curated set of 843 TF binding profiles. The sequence-specific binding of human TFs has been studied using high-throughput SELEX and ChIP sequencing. Motif width is between 7 and 23 (average width 12.7). References:
Text taken from MEME, and partially modified. HOCOMOCO COmprehensive MOdel COllection (HOCOMOCO) provides transcription factor (TF) binding models obtained by careful integration of data from different sources. HOCOMOCO v11 contains 771 non-redundant curated binding models for human TFs. The updated position weight matrices are primarily based on ChIP-Seq data from the GTRD database of the BioUML platform. All the models have been generated using the (ChIPMunk motif discovery tool. Model quality ratings go from A (highest confidence) to D (low confidence). The CORE COLLECTION contains primary binding motifs that robustly represent binding sites across multiple experiments. The models IDs are based on UniProt protein IDs. The FULL collection contains the CORE collection plus all the high-quality alternative and lower-reliability binding models built from limited experimental data. The FULL collection contains models of A/B/C/D quality and includes alternative binding models for a single TF. Motif lenght is between 7 and 25 bp (average width 13.7). References:
Text taken from MEME, and partially modified. The HOCOMOCO COmprehensive MOdel COllection (HOCOMOCO) provides transcription factor (TF) binding models obtained by careful integration of data from different sources. HOCOMOCO v10 contains 531 non-redundant curated binding models for mouse TFs. The updated position weight matrices are primarily based on ChIP-Seq data from the GTRD database of the BioUML platform. All the models have been generated using the (ChIPMunk motif discovery tool. Model quality ratings go from A (highest confidence) to D (low confidence). The models IDs are based on UniProt protein IDs. The CORE COLLECTION contains primary binding motifs that robustly represent binding sites across multiple experiments. The FULL collection contains the CORE collection plus all the high-quality alternative and lower-reliability binding models built from limited experimental data. The FULL collection contains models of A/B/C/D quality and includes alternative binding models for a single TF. Motif lenght is between 7 and 25 bp (average width 13.1). References:
Text taken from MEME, and partially modified. HOCOMOCO COmprehensive MOdel COllection (HOCOMOCO) provides transcription factor (TF) binding models obtained by careful integration of data from different sources. The HOCOMOCO v12 collection provides DNA binding specificity patterns of 949 human transcription factors and 720 mouse orthologs. To make this release, we performed motif discovery in peak sets that originated from 14,183 ChIP-Seq experiments and reads from 2554 HT-SELEX experiments yielding more than 400,000 candidate motifs. The candidate motifs were annotated according to their similarity to known motifs and the hierarchy of DNA-binding domains of the respective transcription factors. Next, the motifs underwent human expert curation to stratify distinct motif subtypes and remove non-informative patterns and common artifacts. Finally, the curated subset of 100,000 motifs was supplied to the automated benchmarking to select the best-performing motifs for each transcription factor. The resulting HOCOMOCO v12 CORE collection contains 1443 verified position weight matrices, including distinct subtypes of DNA binding motifs for particular transcription factors. Motif length is between 7 and 31 in width (average width 15.1). References:
The Isakova 2017 SMiLE-seq Human collection contains a set of 86 binding profiles for human TFs that have been derived using a novel technology, selective microfluidics-based ligand enrichment followed by sequencing (SMiLE-seq). SMiLE-seq is a new technique for the characterization of DNA-binding proteins in a much faster, more accurate and efficient way. The core of SMiLE-seq is a microfluidic platform that involves capillary loading of in vitro-transcribed and -translated bait TFs, and target double-stranded DNA from a pool of random sequences. The transcription factor (TF) is bound to the surface of the microluidic device by antibodies and some fraction of the DNA binds to the TF. The unbound DNA is expelled by washing, so the bound fraction can be measured. Bound DNA is subjected to high-throughput sequencing and a hidden Markov model (HMM)-based TF motif discovery pipeline for de novo identification of DNA-binding specificities and affinities of different families of full-length TFs and TF dimers. Motif width is between 6 and 19 bp (average width 11.8). References:
The Isakova 2017 SMiLE-seq Mouse collection contains a set of 13 binding profiles for mouse TFs that have been derived using a novel technology, selective microfluidics-based ligand enrichment followed by sequencing (SMiLE-seq). SMiLE-seq is a new technique for the characterization of DNA-binding proteins in a much faster, more accurate and efficient way. The core of SMiLE-seq is a microfluidic platform that involves capillary loading of in vitro-transcribed and -translated bait TFs, and target double-stranded DNA from a pool of random sequences. The transcription factor (TF) is bound to the surface of the microluidic device by antibodies and some fraction of the DNA binds to the TF. The unbound DNA is expelled by washing, so the bound fraction can be measured. Bound DNA is subjected to high-throughput sequencing and a hidden Markov model (HMM)-based TF motif discovery pipeline for de novo identification of DNA-binding specificities and affinities of different families of full-length TFs and TF dimers. Motif width is between 6 and 17 bp (average width 12.4). References:
The Isakova 2017 SMiLE-seq Fly collection contains a set of 6 binding profiles for mouse TFs that have been derived using a novel technology, selective microfluidics-based ligand enrichment followed by sequencing (SMiLE-seq). SMiLE-seq is a new technique for the characterization of DNA-binding proteins in a much faster, more accurate and efficient way. The core of SMiLE-seq is a microfluidic platform that involves capillary loading of in vitro-transcribed and -translated bait TFs, and target double-stranded DNA from a pool of random sequences. The transcription factor (TF) is bound to the surface of the microluidic device by antibodies and some fraction of the DNA binds to the TF. The unbound DNA is expelled by washing, so the bound fraction can be measured. Bound DNA is subjected to high-throughput sequencing and a hidden Markov model (HMM)-based TF motif discovery pipeline for de novo identification of DNA-binding specificities and affinities of different families of full-length TFs and TF dimers. Motif width is between 7 and 13 bp (average width 10.8). References:
Text taken from MEME. The Swiss Regulon human and mouse motifs database contains a curated set of 190 mammalian regulatory motifs. Using a combination of data from the JASPAR and TRANSFAC databases, other motifs from the literature, and motifs obtained using their own analysis of ChIP-chip and ChIP-seq data, they curated a data set of 190 position-specific PWMs that represent the binding specificities of ~340 TFs in both human and mouse. The curation included reducing redundancy by fusing PWMs that are similar, have dominantly overlapping binding sites or are associated with TFs sharing highly similar DNA-binding domains. PWMs were also refined by iteratively performing TFBS predictions in all proximal promoters genome wide. Motif width is between 4 and 30 bp (average width 12.5). References:
Text taken from MEME. UniPROBE Mouse provides 386 transcription factor (TF) motifs from protein-binding microarrays downloaded from UniPROBE. Motif width is between 13 and 23bp (average width 16.3). References:
Text taken from MEME, and partially modified. UniPROBE Worm provides 36 motifs of c. elegans homeodomain proteins UniPROBE. Motif width is between 14 and 17 bp (average width 15.4). References:
Text taken from MEME. The Swiss Regulon human and mouse motifs database contains a curated set of 158 S. cerevisiae regulatory motifs based on ChIP-chip and in vitro assays. Motif width is between 5 and 19 bp (average width 10.1). References:
Text taken from JASPAR DOWNLOAD, and partially modified. The JASPAR CORE database contains a curated non-redundant set of 143 profiles derived from published collections of experimentally defined transcription factor binding sites for insects. The new and updated profiles are mainly derived from published chromatin immunoprecipitation-seq experimental datasets. Motif lenght is between between 5 and 21 bp (average width 8.5). References:
Text taken from MEME. The OnTheFly database is a systematic collection of Drosophila melanogaster transcription factors and their DNA-binding sites. All Transcription Factors (TFs) predicted in the Drosophila melanogaster genome have been annotated, and the known preferred DNA binding sites of the TFs based on the B1H, DNaseI and SELEX experimental methods have been collected. OnTheFly houses DNA recognition motifs for 387 different genes encoding TFs (>50% of the Drosophila melanogaster genes encoding TFs). The collection provides 608 motifs whose length is between 4 and 23 bp (average width 9.3). References:
Text taken from JASPAR DOWNLOAD, and partially modified. The JASPAR CORE database provides a curated non-redundant set of 530 profiles derived from published collections of experimentally defined transcription factor binding sites for plants. This release included 164 new curated profiles. Motif lenght is between between 5 and 30 bp (average width 12.6). References:
Text taken from MEME, and modified. The Arabidopsis DAPv1 O'Malley2016 collection contains transcription factor (TF) binding site motifs derived from DNA affinity purification sequencing (DAP-seq), a high-throughput assay that uses in-vitro-expressed TF to interrogate naked genomic DNA fragments to establish binding locations (peaks) and sequence motifs. The goal of this study was to develop robust methods to build comprehensive cistrome and epicistrome maps in Arabidopsis. The position weight matrices from experiments performed on leaf genomic DNA libraries and amplified DNA libraries (methylation-free) were provided by the authors. This collection includes 872 motifs whose length is between 5 and 30 bp (average width 15.2). References:
References:
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 8 and 8 (average width 8.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 8 and 8 (average width 8.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 11 and 11 (average width 11.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 9 and 9 (average width 9.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 208. Motif width is between 5 and 25 (average width 10.4).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 229. Motif width is between 5 and 21 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 12. Motif width is between 7 and 21 (average width 10.4).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 612. Motif width is between 6 and 25 (average width 11.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 13. Motif width is between 5 and 21 (average width 10.2).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 45. Motif width is between 5 and 21 (average width 9.4).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 54. Motif width is between 7 and 21 (average width 11.4).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 474. Motif width is between 7 and 30 (average width 11.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 231. Motif width is between 5 and 24 (average width 10.1).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 4. Motif width is between 7 and 14 (average width 10.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 243. Motif width is between 5 and 21 (average width 10.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 8 and 8 (average width 8.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 287. Motif width is between 7 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 309. Motif width is between 7 and 21 (average width 9.8).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 8 and 8 (average width 8.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 43. Motif width is between 5 and 12 (average width 9.3).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 53. Motif width is between 5 and 21 (average width 10.4).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 54. Motif width is between 5 and 21 (average width 9.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 59. Motif width is between 5 and 21 (average width 9.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 56. Motif width is between 5 and 21 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 53. Motif width is between 5 and 21 (average width 9.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 58. Motif width is between 5 and 21 (average width 9.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 55. Motif width is between 5 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 55. Motif width is between 5 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 57. Motif width is between 5 and 21 (average width 9.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 8 and 8 (average width 8.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 10 and 10 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 8 and 8 (average width 8.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 13 and 13 (average width 13.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 3. Motif width is between 8 and 8 (average width 8.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 5. Motif width is between 10 and 21 (average width 14.4).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 55. Motif width is between 5 and 21 (average width 10.1).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 181. Motif width is between 5 and 20 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 584. Motif width is between 6 and 30 (average width 11.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 52. Motif width is between 5 and 21 (average width 9.8).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 193. Motif width is between 7 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 272. Motif width is between 5 and 25 (average width 11.2).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 3. Motif width is between 8 and 10 (average width 9.3).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 106. Motif width is between 6 and 21 (average width 10.1).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 8 and 8 (average width 8.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 180. Motif width is between 7 and 21 (average width 9.8).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 209. Motif width is between 6 and 21 (average width 9.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 269. Motif width is between 6 and 21 (average width 9.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 171. Motif width is between 6 and 21 (average width 9.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 213. Motif width is between 6 and 21 (average width 9.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 622. Motif width is between 6 and 25 (average width 11.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 33. Motif width is between 5 and 21 (average width 10.3).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 30. Motif width is between 5 and 21 (average width 10.4).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 64. Motif width is between 5 and 21 (average width 10.3).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 24. Motif width is between 5 and 21 (average width 10.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 23. Motif width is between 5 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 30. Motif width is between 5 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 27. Motif width is between 5 and 21 (average width 9.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 584. Motif width is between 6 and 25 (average width 11.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 191. Motif width is between 7 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 249. Motif width is between 5 and 25 (average width 10.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 15. Motif width is between 7 and 18 (average width 10.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 586. Motif width is between 7 and 25 (average width 11.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 66. Motif width is between 5 and 14 (average width 9.1).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 6. Motif width is between 8 and 11 (average width 9.2).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 6. Motif width is between 8 and 11 (average width 9.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 7. Motif width is between 8 and 21 (average width 11.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 232. Motif width is between 6 and 25 (average width 11.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 153. Motif width is between 5 and 25 (average width 10.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 152. Motif width is between 5 and 25 (average width 10.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 56. Motif width is between 5 and 21 (average width 10.2).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 46. Motif width is between 7 and 21 (average width 10.3).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 7. Motif width is between 8 and 21 (average width 11.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 55. Motif width is between 5 and 21 (average width 9.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 78. Motif width is between 5 and 21 (average width 9.8).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 2. Motif width is between 9 and 10 (average width 9.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 17. Motif width is between 5 and 21 (average width 9.4).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 9 and 9 (average width 9.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 67. Motif width is between 5 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 13. Motif width is between 5 and 21 (average width 9.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 8 and 8 (average width 8.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 8 and 8 (average width 8.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 8 and 8 (average width 8.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 208. Motif width is between 7 and 21 (average width 10.1).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 205. Motif width is between 5 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 205. Motif width is between 6 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 8 and 8 (average width 8.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 552. Motif width is between 7 and 25 (average width 11.2).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 228. Motif width is between 5 and 21 (average width 10.3).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 312. Motif width is between 6 and 25 (average width 11.3).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 27. Motif width is between 5 and 21 (average width 10.3).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 41. Motif width is between 7 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 28. Motif width is between 7 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 389. Motif width is between 6 and 24 (average width 11.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 8 and 8 (average width 8.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 300. Motif width is between 5 and 25 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 322. Motif width is between 5 and 25 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 299. Motif width is between 5 and 25 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 361. Motif width is between 5 and 25 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 301. Motif width is between 6 and 25 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 289. Motif width is between 5 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 305. Motif width is between 5 and 25 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 312. Motif width is between 5 and 25 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 284. Motif width is between 5 and 25 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 308. Motif width is between 5 and 25 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 315. Motif width is between 5 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 321. Motif width is between 6 and 25 (average width 10.1).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 10 and 10 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 339. Motif width is between 6 and 24 (average width 11.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 2. Motif width is between 8 and 11 (average width 9.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 15 and 15 (average width 15.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 7. Motif width is between 8 and 21 (average width 14.1).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 7. Motif width is between 8 and 21 (average width 14.1).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 3. Motif width is between 8 and 21 (average width 13.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 557. Motif width is between 6 and 25 (average width 11.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 319. Motif width is between 7 and 24 (average width 11.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 355. Motif width is between 6 and 25 (average width 11.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 2. Motif width is between 7 and 9 (average width 8.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 76. Motif width is between 5 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 69. Motif width is between 5 and 21 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 68. Motif width is between 5 and 21 (average width 9.8).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 414. Motif width is between 7 and 25 (average width 11.3).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 530. Motif width is between 7 and 30 (average width 11.4).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 11 and 11 (average width 11.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 237. Motif width is between 7 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 628. Motif width is between 6 and 25 (average width 11.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 21 and 21 (average width 21.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 9 and 9 (average width 9.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 236. Motif width is between 5 and 22 (average width 10.8).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 10 and 10 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 11. Motif width is between 8 and 21 (average width 10.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 8 and 8 (average width 8.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 42. Motif width is between 6 and 21 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 733. Motif width is between 6 and 25 (average width 11.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 6. Motif width is between 8 and 20 (average width 12.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 72. Motif width is between 5 and 25 (average width 10.8).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 10 and 10 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 167. Motif width is between 5 and 25 (average width 10.8).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 8 and 8 (average width 8.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 48. Motif width is between 5 and 24 (average width 10.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 55. Motif width is between 5 and 21 (average width 10.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 49. Motif width is between 5 and 21 (average width 9.8).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 15. Motif width is between 6 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 60. Motif width is between 5 and 21 (average width 10.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 11 and 11 (average width 11.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 118. Motif width is between 7 and 21 (average width 10.1).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 26. Motif width is between 5 and 21 (average width 9.2).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 243. Motif width is between 5 and 25 (average width 10.8).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 181. Motif width is between 7 and 21 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 578. Motif width is between 6 and 30 (average width 11.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 626. Motif width is between 6 and 30 (average width 11.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 406. Motif width is between 6 and 30 (average width 11.4).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 64. Motif width is between 5 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 6. Motif width is between 5 and 21 (average width 10.2).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 219. Motif width is between 7 and 21 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 240. Motif width is between 7 and 21 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 10 and 10 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 184. Motif width is between 7 and 21 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 9. Motif width is between 5 and 21 (average width 9.3).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 340. Motif width is between 7 and 30 (average width 11.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 67. Motif width is between 7 and 16 (average width 9.8).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 449. Motif width is between 7 and 25 (average width 11.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 13. Motif width is between 8 and 21 (average width 11.4).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 54. Motif width is between 5 and 21 (average width 9.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 54. Motif width is between 5 and 21 (average width 9.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 204. Motif width is between 7 and 21 (average width 9.8).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 2. Motif width is between 8 and 10 (average width 9.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 564. Motif width is between 6 and 25 (average width 11.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 6. Motif width is between 8 and 13 (average width 10.3).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 24. Motif width is between 5 and 21 (average width 9.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 718. Motif width is between 6 and 30 (average width 10.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 70. Motif width is between 5 and 14 (average width 9.2).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 44. Motif width is between 5 and 21 (average width 9.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 49. Motif width is between 5 and 21 (average width 9.4).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 372. Motif width is between 6 and 25 (average width 11.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 5. Motif width is between 8 and 21 (average width 11.2).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 226. Motif width is between 5 and 21 (average width 10.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 74. Motif width is between 5 and 21 (average width 9.8).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 203. Motif width is between 6 and 25 (average width 10.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 57. Motif width is between 5 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 2. Motif width is between 8 and 8 (average width 8.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 130. Motif width is between 5 and 21 (average width 9.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 111. Motif width is between 5 and 21 (average width 9.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 113. Motif width is between 5 and 21 (average width 9.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 7 and 7 (average width 7.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 19. Motif width is between 7 and 21 (average width 11.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 423. Motif width is between 7 and 30 (average width 11.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 96. Motif width is between 5 and 25 (average width 11.1).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 7 and 7 (average width 7.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 366. Motif width is between 7 and 25 (average width 11.4).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 548. Motif width is between 7 and 30 (average width 11.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 194. Motif width is between 7 and 21 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 203. Motif width is between 7 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 510. Motif width is between 7 and 25 (average width 11.3).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 17. Motif width is between 8 and 21 (average width 10.4).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 17. Motif width is between 8 and 21 (average width 10.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 29. Motif width is between 8 and 21 (average width 10.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 358. Motif width is between 7 and 25 (average width 11.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 33. Motif width is between 8 and 12 (average width 8.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 9 and 9 (average width 9.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 625. Motif width is between 6 and 30 (average width 11.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 42. Motif width is between 6 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 6. Motif width is between 8 and 10 (average width 8.8).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 235. Motif width is between 5 and 21 (average width 10.4).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 59. Motif width is between 5 and 21 (average width 10.1).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 59. Motif width is between 5 and 21 (average width 10.1).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 7 and 7 (average width 7.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 5. Motif width is between 8 and 12 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 8. Motif width is between 9 and 21 (average width 15.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 9 and 9 (average width 9.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 46. Motif width is between 5 and 21 (average width 9.4).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 8. Motif width is between 5 and 21 (average width 10.8).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 10 and 10 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 86. Motif width is between 7 and 21 (average width 9.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 22. Motif width is between 5 and 21 (average width 9.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 117. Motif width is between 7 and 21 (average width 10.1).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 7. Motif width is between 5 and 15 (average width 10.3).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 6. Motif width is between 5 and 15 (average width 9.8).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 9. Motif width is between 5 and 15 (average width 10.1).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 6. Motif width is between 9 and 15 (average width 10.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 9 and 9 (average width 9.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 31. Motif width is between 5 and 21 (average width 10.8).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 29. Motif width is between 5 and 21 (average width 9.4).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 4. Motif width is between 7 and 16 (average width 10.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 12. Motif width is between 8 and 8 (average width 8.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 12. Motif width is between 8 and 8 (average width 8.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 19. Motif width is between 8 and 9 (average width 8.1).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 12. Motif width is between 8 and 8 (average width 8.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 12. Motif width is between 8 and 8 (average width 8.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 11. Motif width is between 8 and 8 (average width 8.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 10 and 10 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 8 and 8 (average width 8.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 13. Motif width is between 5 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 8. Motif width is between 6 and 21 (average width 13.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 10 and 10 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 83. Motif width is between 5 and 21 (average width 9.4).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 27. Motif width is between 7 and 18 (average width 9.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 606. Motif width is between 6 and 25 (average width 11.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 233. Motif width is between 7 and 21 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 14. Motif width is between 5 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 74. Motif width is between 6 and 20 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 408. Motif width is between 6 and 22 (average width 11.3).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 212. Motif width is between 7 and 21 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 516. Motif width is between 6 and 24 (average width 11.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 10. Motif width is between 5 and 21 (average width 9.4).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 7. Motif width is between 8 and 21 (average width 10.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 54. Motif width is between 5 and 21 (average width 9.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 620. Motif width is between 6 and 30 (average width 11.3).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 21. Motif width is between 5 and 21 (average width 9.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 218. Motif width is between 7 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 129. Motif width is between 5 and 24 (average width 11.3).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 63. Motif width is between 5 and 21 (average width 11.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 218. Motif width is between 5 and 29 (average width 11.3).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 91. Motif width is between 5 and 23 (average width 11.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 143. Motif width is between 5 and 24 (average width 11.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 152. Motif width is between 5 and 24 (average width 11.1).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 9. Motif width is between 8 and 15 (average width 11.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 2. Motif width is between 10 and 15 (average width 12.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 10 and 10 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 93. Motif width is between 6 and 22 (average width 10.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 111. Motif width is between 6 and 22 (average width 10.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 13. Motif width is between 5 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 12. Motif width is between 5 and 21 (average width 11.3).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 14. Motif width is between 5 and 21 (average width 10.4).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 14. Motif width is between 5 and 21 (average width 10.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 15. Motif width is between 5 and 21 (average width 10.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 57. Motif width is between 5 and 21 (average width 9.8).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 95. Motif width is between 7 and 21 (average width 10.1).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 10. Motif width is between 5 and 21 (average width 10.3).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 15. Motif width is between 8 and 21 (average width 11.8).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 226. Motif width is between 7 and 24 (average width 11.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 210. Motif width is between 7 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 10 and 10 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 290. Motif width is between 7 and 30 (average width 11.4).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 15. Motif width is between 8 and 21 (average width 11.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 8. Motif width is between 5 and 21 (average width 11.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 55. Motif width is between 5 and 21 (average width 9.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 221. Motif width is between 6 and 25 (average width 11.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 490. Motif width is between 6 and 30 (average width 11.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 432. Motif width is between 7 and 25 (average width 11.3).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 547. Motif width is between 6 and 25 (average width 11.3).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 268. Motif width is between 6 and 25 (average width 11.8).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 4. Motif width is between 9 and 21 (average width 12.2).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 538. Motif width is between 6 and 30 (average width 11.2).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 3. Motif width is between 8 and 9 (average width 8.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 4. Motif width is between 9 and 21 (average width 12.2).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 4. Motif width is between 8 and 15 (average width 9.8).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 3. Motif width is between 8 and 8 (average width 8.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 78. Motif width is between 5 and 21 (average width 9.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 2. Motif width is between 8 and 8 (average width 8.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 11. Motif width is between 5 and 21 (average width 10.2).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 39. Motif width is between 5 and 17 (average width 10.2).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 69. Motif width is between 5 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 62. Motif width is between 5 and 21 (average width 9.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 69. Motif width is between 5 and 21 (average width 9.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 13. Motif width is between 7 and 11 (average width 9.8).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 48. Motif width is between 7 and 12 (average width 9.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 53. Motif width is between 5 and 21 (average width 9.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 48. Motif width is between 7 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 42. Motif width is between 5 and 21 (average width 9.8).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 92. Motif width is between 7 and 25 (average width 11.3).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 9 and 9 (average width 9.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 8 and 8 (average width 8.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 9. Motif width is between 8 and 20 (average width 10.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 1. Motif width is between 7 and 7 (average width 7.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 56. Motif width is between 5 and 21 (average width 9.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 315. Motif width is between 6 and 24 (average width 11.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 526. Motif width is between 6 and 25 (average width 11.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 44. Motif width is between 5 and 21 (average width 10.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 14. Motif width is between 7 and 21 (average width 10.3).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 53. Motif width is between 5 and 21 (average width 10.9).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 46. Motif width is between 5 and 16 (average width 9.3).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 67. Motif width is between 6 and 21 (average width 9.7).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 216. Motif width is between 7 and 25 (average width 11.5).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 189. Motif width is between 7 and 21 (average width 10.0).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 6. Motif width is between 8 and 11 (average width 9.3).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 105. Motif width is between 7 and 21 (average width 10.1).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 467. Motif width is between 6 and 30 (average width 11.6).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 481. Motif width is between 6 and 25 (average width 11.4).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 33. Motif width is between 5 and 21 (average width 10.1).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 206. Motif width is between 5 and 21 (average width 9.8).
Text taken from MEME, and partially modified. Direct and inferred motifs for Acipenser baerii from the CisBP (Catalog of Inferred Sequence Binding Preferences) database. To reduce redundancy, for each transcription factor (TF) in this species that has a CisBP motif, we selected a single motif according to the following precedence rules. We chose the direct motif if there is one, otherwise we chose the inferred motif with the highest DNA binding domain (DBD) similarity (according to CisBP) to a TF in another species that has a direct motif. If there is more than one direct motif or inferred motif with the highest DBD similarity, we chose among them according to their provenance (CisBP's "Motif_Type" attribute) in the following order: ChIP-seq, HocoMoco, DeBoer11, PBM, SELEX, B1H, High-throughput Selex CAGE, PBM:CSA:DIP-chip, ChIP-chip, COMPILED, DNaseI footprinting. We then linked each motif thus determined to a single TF in the CisBP database, following the same precedence rules. Total number of motifs: 64. Motif width is between 5 and 24 (average width 11.4).
|


