TSS assembly pipeline for At_EPDnew_004
Introduction
This document provides a technical description of the transcription start site assembly pipeline that was used to generate EPDnew version 004 for A. thaliana.
Source Data
Promoter collection:
| Name | Genome Assembly | Promoters | Genes | PMID | Access data | ||
|---|---|---|---|---|---|---|---|
| TAIR10 Genes | Feb 2011 TAIR10/araTha1 | 31615 | 27149 | 26201819 | SOURCE | DOC | DATA |
Experimental data:
| Name | Type | Samples | Tags | PMID | Access data | ||
|---|---|---|---|---|---|---|---|
| Morton et al., 2014 | PEAT | 1 | 22,578,668 | 25035402 | SOURCE | DOC | DATA |
| Cumbie et al., 2015 | NanoCAGE-XL | 3 | 226,177,104 | 26268438 | SOURCE | DOC | DATA |
| Tokizawa et al., 2017 | CAGE and OligoCap | 2 | 178,605,048 | 28214361 | SOURCE | DOC | DATA |
| Ushijima et al., 2017 | CAGE | 7 | 552,864,502 | 29129375 | SOURCE | DOC | DATA |
Assembly pipeline overview
|
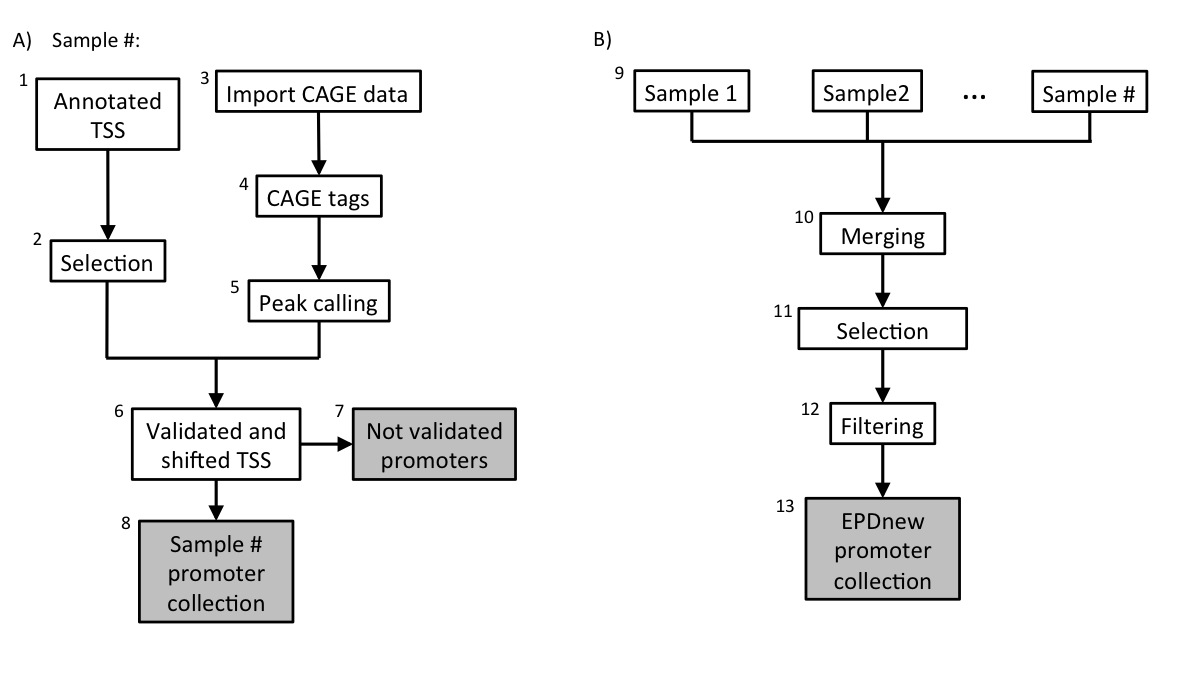
Description of procedures and intermediate data files
1. Download annotated TSS
Primary annotation data was downloaded from TAIR the 06-02-2015.
Genes annotations downloaded from TAIR did not contain direct links to RefSeq ID. For this reason, RefSeq ID has been parsed from NCBI RefSeq files.
A total of 31615 promoters were selected.2. TAIR10 TSS collection
The TAIR10 TSS collection is stored as a tab-deliminated text file conforming to the SGA format under the name:
-
arabidopsisTair10Genes.sga
- NCBI/RefSeq chromosome id
- "TSS"
- position
- strand ("+" or "-")
- "1"
- TAIR ID
3. Import CAGE data
Data was imported from the lab web page or GEO in BAM or SRA file formats. Please refer to the "Source Data" table at the beginning of this document for the links to raw data archives. A detailed guide on how to import, map and convert these samples can be found in the corresponding "MGA doc" files.
5. mRNA 5' tags peak calling
For the 12 samples present, peak calling has been carried out using ChIP-Peak on-line tool with the following parameters:
- Window width = 1
- Vicinity range = 200
- Peak refine = Y
- Count cutoff = 9999999
- Threshold = 5
6. TSS validation and shifting
The 12 samples used were then individually processed in a pipeline aiming at validating transcription start sites with mRNA peaks. A TAIR10 TSS was experimentally confirmed if a CAGE peak lied in a window of 100 bp around it. The validated TSS was then shifted to the nearest base with the higher tag density.
7. Promoter collections
Each sample in the dataset was used to generate a separate promoter collection. Potentially, the same transcript could be validated by multiple samples and it could have different start sites in different samples. To avoid redundancy, the individual collections were used as input for an additional step in the analysis (Assembly pipeline part B).
8. Quality controls
Each promoter collection was then checked for the presence of core promoter elements (TATA-box and Inr) in the expected positions.
9. Merging sample-specific collections and further TSS selection
All sample-specific promoter collections were merged into a unique file and further analyzed. Transcripts validated by multiple samples could potentially have the TSS set on a broader region rather than a single position. To avoid such inconsistency, for each transcript we selected the position that was validated by the largest number of samples as the true TSS.
10. Filtering
TSSs that mapped close to other TSSs that belonged to the same gene (100-bp window) were merged into a unique promoter following the same rule: the promoter that was validated by the highest number of samples was kept.
11. Final EPDnew collection
The 22703
experimentally validated promoters were stored in the
EPDnew database, which can be downloaded from our ftp
site. Scientists are welcome to use our other tools
ChIP-Seq
(for correlation analysis) and
SSA
(for motif analysis around promoters) to analyze the
EPDnew database.