TSS assembly pipeline for Hs_EPDnew_004
Introduction
This document provides a technical description of the transcription start site assembly pipeline that was used to generate EPDnew version 004 for H. sapiens.Source Data
Promoter collection:
| Name | Genome Assembly | Promoters | Genes | PMID | Access data | ||
|---|---|---|---|---|---|---|---|
| UCSC Known Genes | Feb 2009 GRCh37/hg19 | 28210 | 18636 | 26590259 | SOURCE | DOC | DATA |
Experimental data:
| Name | Type | Samples | Tags | PMID | Access data | ||
|---|---|---|---|---|---|---|---|
| FANTOM5 | CAGE | 941 | 18,244,201,540 | 24670764 | SOURCE | DOC | DATA |
| ENCODE | CAGE | 145 | 7,134,200,060 | 22955620 | SOURCE | DOC | DATA |
Assembly pipeline overview
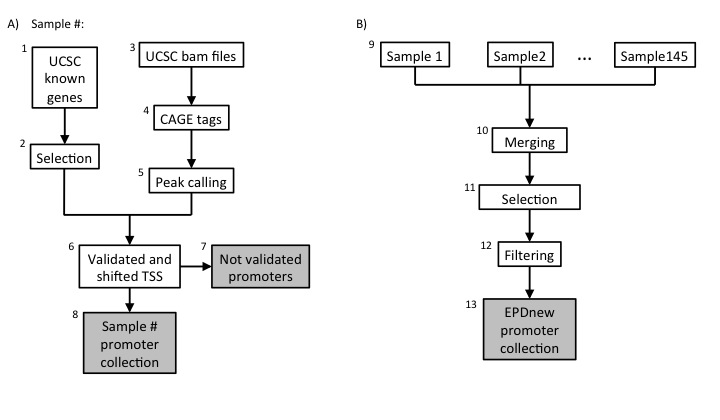
|
Description of procedures and intermediate data files
1. UCSC Download
Data was downloaded from UCSC Table Browser (11-03-2014) selecting the following attributes:- hg19.knownGene
- hg19.ensemblSource
- hg19.kgXref
- hg19.knownToEnsembl
- hg19.refSeqStatus
- hg19.spMrna
- Transcripts of protein coding genes only (Ensembl annotation)
- Transcripts must have a RefSeq protein ID
- Transcripts must have a non "n/a" RefSeq status
Transcripts with the same TSS position were merged under a common ID. As a consequence of this the total number of TSS in the list was 28210.
2. UCSC TSS collection
The UCSC TSS collection is stored as a tab-deliminated text file conforming to the SGA format under the name:-
ucsc_promoter_list.fps
- NCBI/RefSeq chromosome id
- "TSS"
- position
- strand ("+" or "-")
- "1"
- TranscriptID..GeneName.
3. Data import from ENCODE and FANTOM5 CAGE
CAGE Tag Data were downloaded from UCSC ftp-site and FANTOM5 http-site (see links above). The source files are in bam format. The complete list of files can be found here for ENCODE and here for FANTOM5. Bam files were converted into bed files with bamToBed program. Files were kept and analysed individually.4. CAGE tags
The compressed versions of these files are available from the MGA archive (see links above).5. mRNA 5' tags peak calling
Peak calling for each individual CAGE data file has been carried out using ChIP-Peak on-line tool with the following parameters:- Window width = 200
- Vicinity range = 200
- Peak refine = N
- Count cutoff = 9999999
- Threshold = 3
6. TSS validation and shifting
Each sample in the collection (mRNA peaks and UCSC TSS) was then processed in a pipeline aiming at validating transcription start sites with mRNA peaks. An UCSC TSS was experimentally confirmed if a CAGE peak lied in a window of 500 bp around it and if it had a maximum high of at least 3 tags. The validated TSS was then shifted to the nearest base with the higher tag density.7. UCSC not-validated TSS
The total number (summing up all samples) of non experimentally validated TSS was around 3000.8. Promoter collection for each sample
Each sample in the dataset was used to generate a separate promoter collection. Potentially, the same transcript could be validated by multiple samples and it could have different start sites in different samples. To avoid redundancy, the individual collections were used as input for an additional step in the analysis (Assembly pipeline part B).9. Merging collections and second TSS selection
The 1K promoter collections were merged into a unique file and further analysed. The promoter of a transcript was mantained in the list only if validated by at least two samples. Transcript validated by multiple samples could potentially have the TSS set on a broader region and not to single position. To avoid such inconsistency, for each transcript we selected the position that was validated by the larger number of samples as the true TSS.10. Filtering
Transcription Start Sites that mapped closed to other TSS that belonged to the same gene (500 bp window) were merged into a unique promoter following the same rule: the promoter that was validated by the higher number of samples was kept.10. Final EPDnew collection
The 25503 experimentally validated promoter were stored in the EPDnew database that can be downloaded from our ftp site. Scientist are wellcome to use our other tools ChIP-Seq (for correlation analysis) and SSA (for motifs analysis around promoters) to analyse EPDnew database.Last update October 2019