ChIP-Cor quick guide
This page is intended as a very quick guide to ChIP-Cor, one of the tools of the ChIP-Seq analysis tools. Comprehensive instructions can be found here.
Overview
ChIP-Cor is a tool that correlate two features (called Reference and Target) and calculate the density distribution of the Target feature around the Reference feature. For example, it can be used to visualize the distribution of the histone marks H3K4me3 (Target feature) around promoters (Reference feature).
How-to
1. Select the Reference Feature
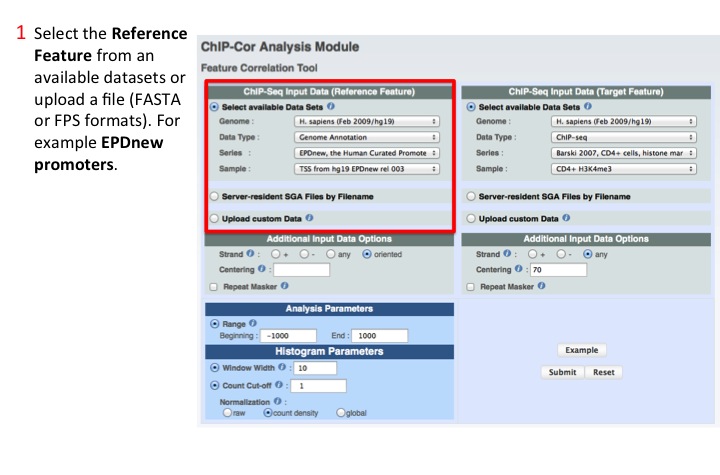
Input can be one of the available data sets or a user provided file in several formats. In this example we selected human transcription start sites (TSS) from EPDnew.
2. Adjust Reference parameters
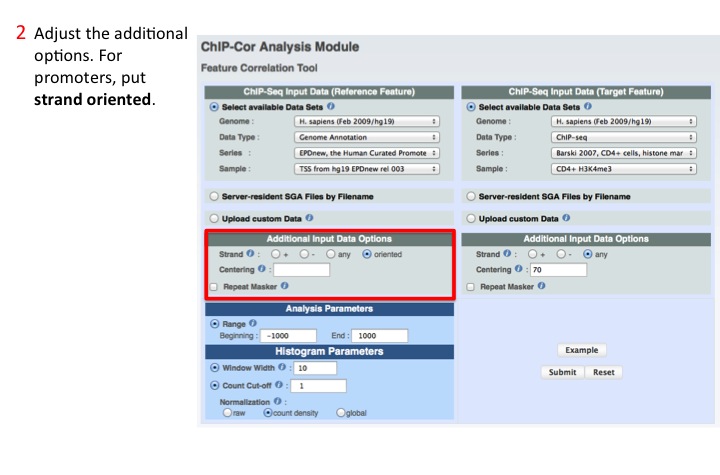
Possible parameters are: Strand, Centering and Repeat Masker. Here, TSS are oriented feature (the region before them is the promoter, whereas after is the gene body).
3. Select Target Feature
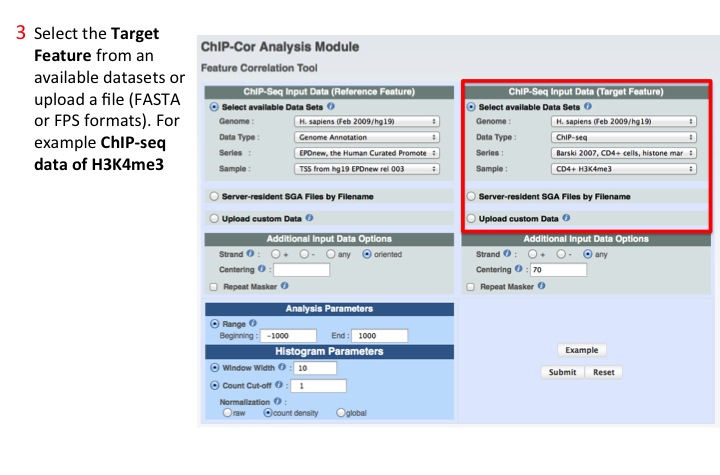
As for the Reference, input can be one of the available data sets or a user provided file in several formats. In this example we selected the histone variants H3K4me3.
4. Adjust Target parameters
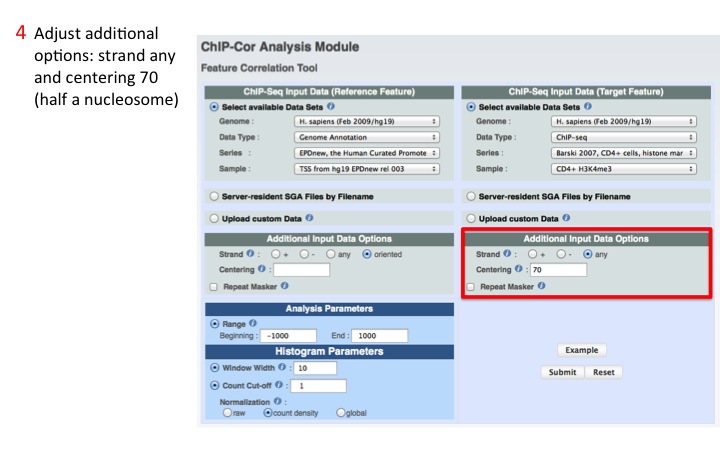
Possible parameters are: Strand, Centering and Repeat Masker. Here, we select any strand and we centering of half a nucleosome (70 bp).
5. Adjust additional parameters
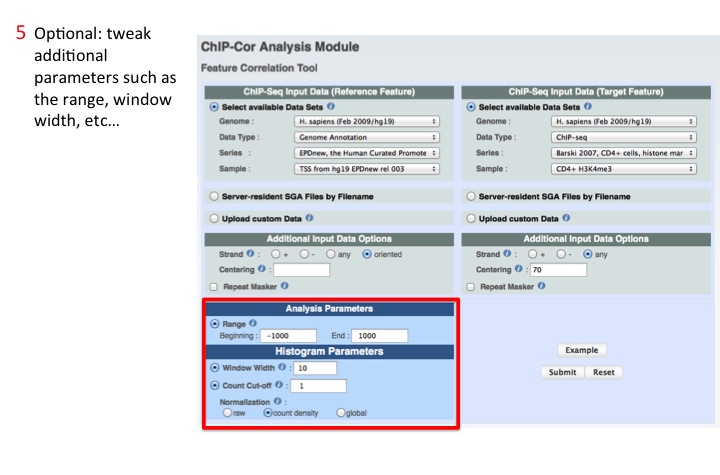
Possible parameters are: Range of the Region, Window widht and count cut-off.
6. Get the results
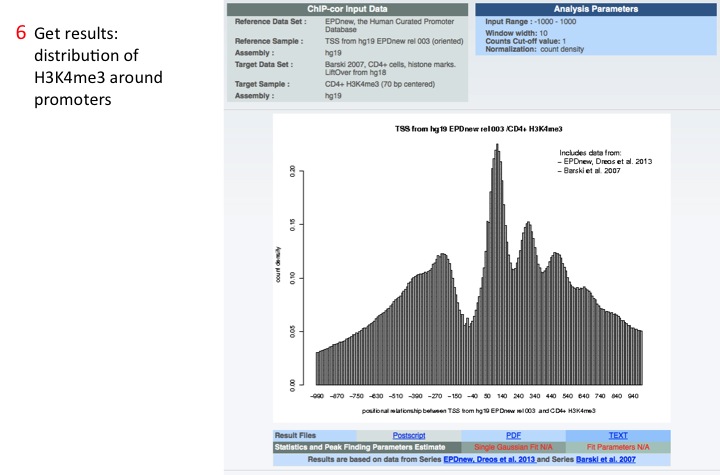
ChIP-Cor plots the distribution of the Target feature around the Reference feature.