OProf quick guide
This page is intended as a very quick guide to OProf, one of the tools of the Signal Search Analysis Server. Comprehensive instructions can be found here.
Overview
OProf is a tool that scans a set of DNA sequences aligned with respect to a functional site (e.g. a transcription start site) to determine the frequency of a motif in a region around the functional site. For example it can show the distribution of the TATA-box around promoters.
How-to
1. Select the input
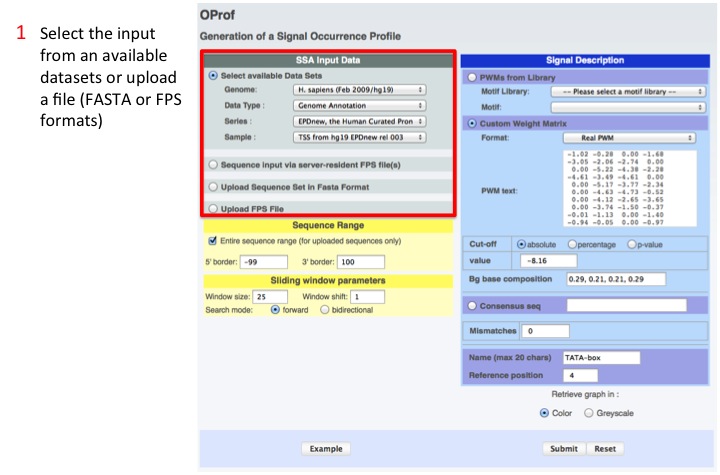
Inputs can be one of the available data sets, a user provided FASTA file or a users' FPS file.
2. Select the motif
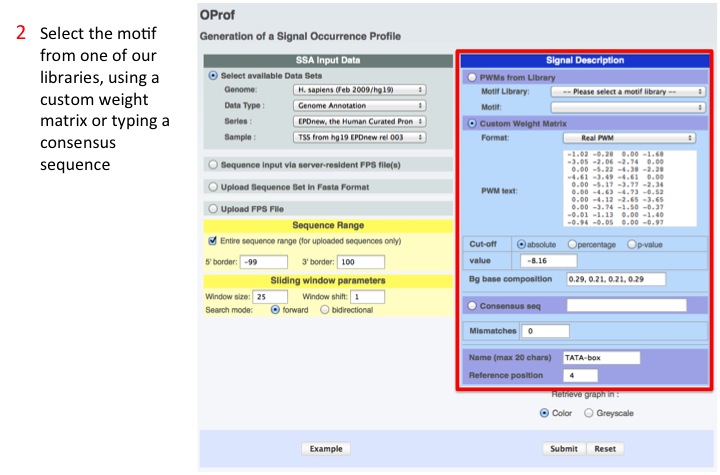
A motif can be selected from the available libraries, a user provided Position Weight Matrix or a consensus sequence.
3. Select additional parameters
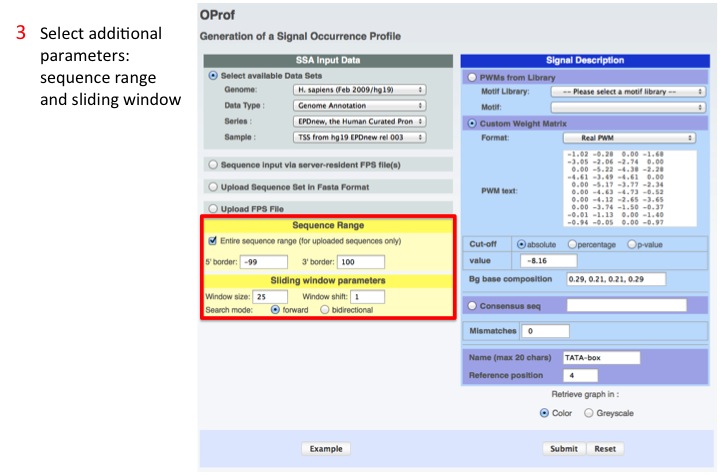
Sequence range: is the range around the functional site that OProf scans for the motif
Sliding Window: is the window width and shift used by OProf when scanning for the motif
4. Get the results
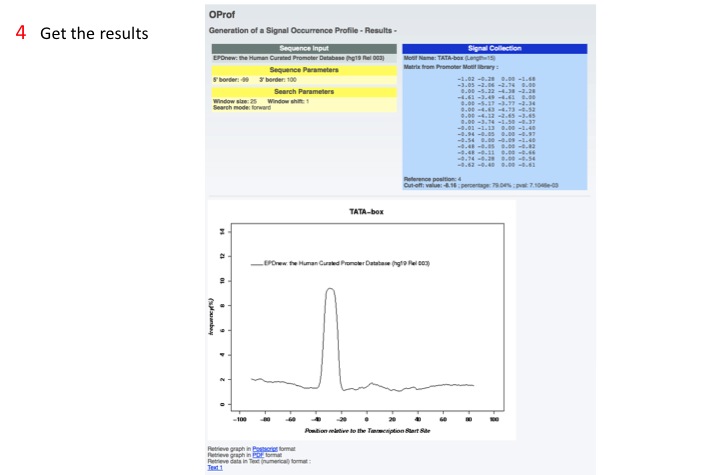
OProf shows the distribution of the motif around the functional site (in this example the TATA-box in EPDnew promoters, alligned by the TSS).