Lausanne, 27 March - 31 March 2017
| Exercise | Publication details | Relevant figure |
|---|---|---|
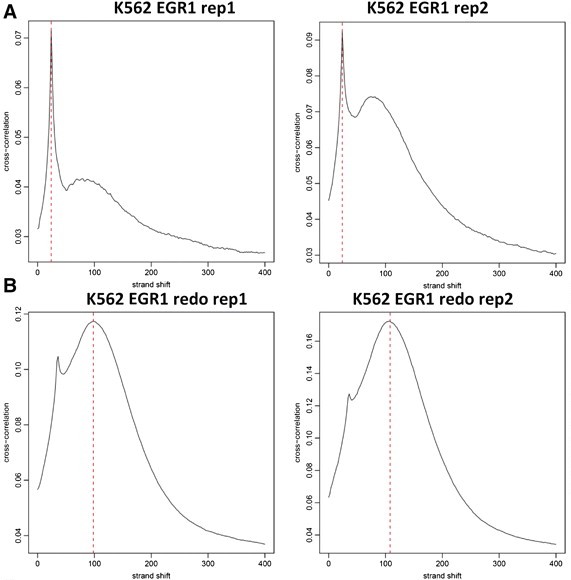
| Cross-correlaiton analysis | Landt et al. 2012, ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Research 22, 1813-1831. PMID 22955991 |  |
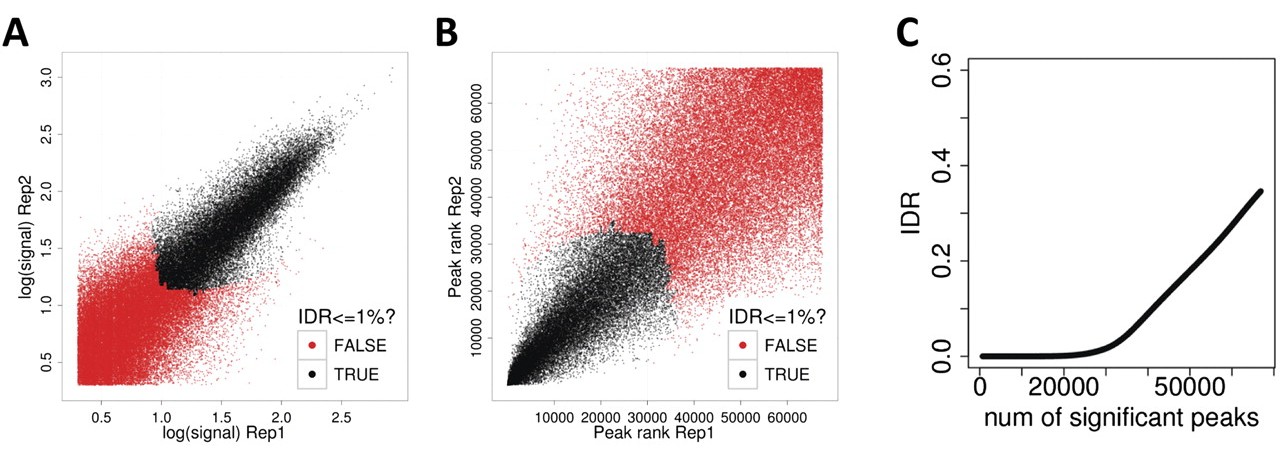
| Irreproducible Discovery Rate (IDR) | Landt et al. 2012, ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Research 22, 1813-1831. PMID 22955991 | 
|
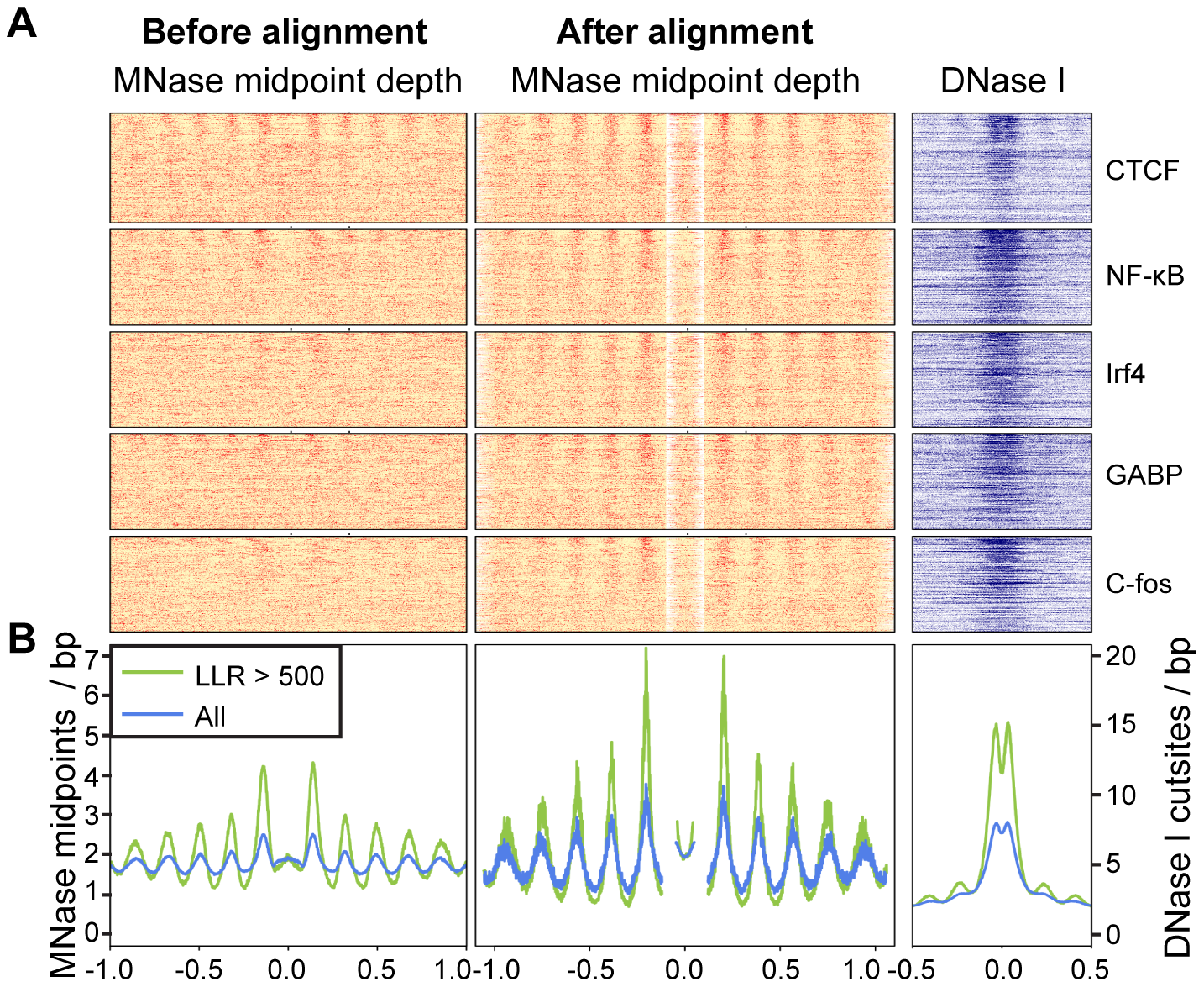
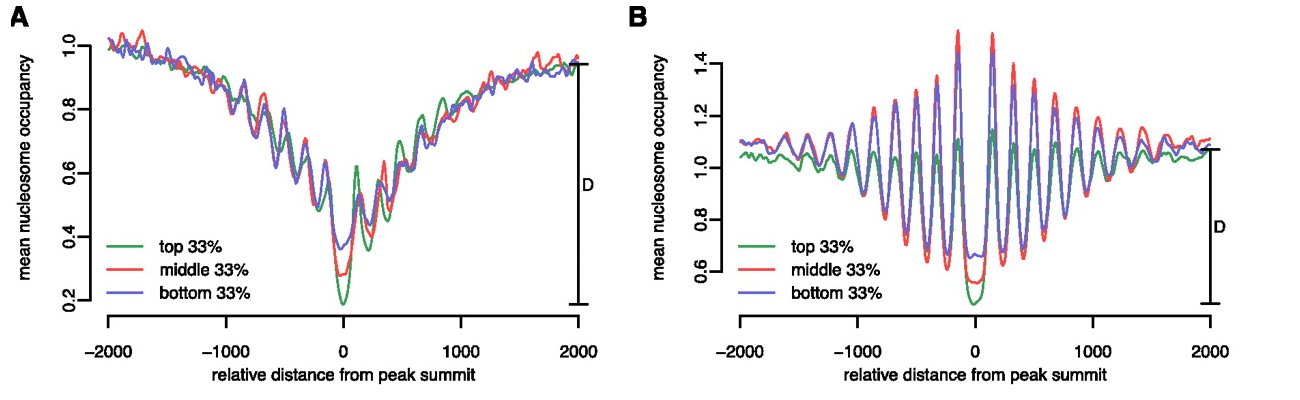
| Alignment of MNase tags around NFKB sites | Gaffney et al. 2012, Controls of nucleosome positioning in the human genome. PLoS Genetics. 8: e1003036. PMID 23166509 |  |
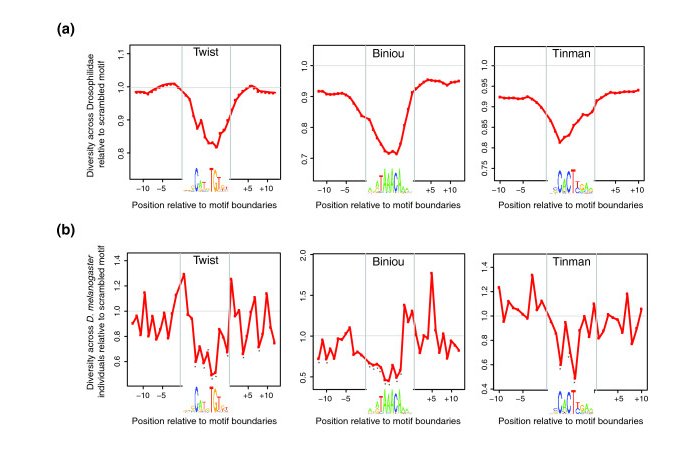
| Variation in Drosophila transcription factor binding sites | Spivakov et al. 2012. Analysis of variation at transcription factor binding sites in Drosophila and humans. Genome Biol. 13:R49. PMID 22950968 |  |
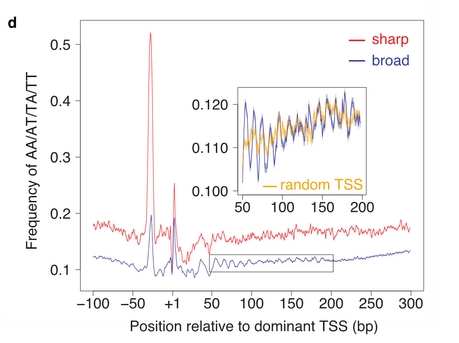
| Epigenetic variation across promoter types | Nozaki et al. 2011. Tight associations between transcription promoter type and epigenetic variation in histone positioning and modification BMC Genomics, 12:416 PMID 21846408 |  |
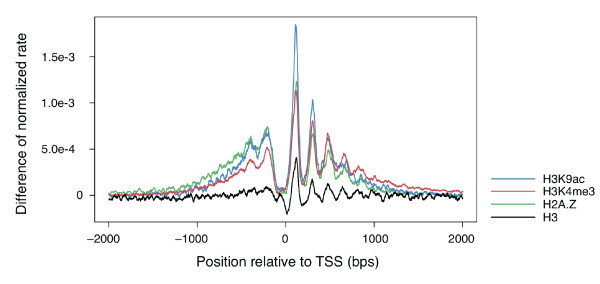
| Nucleosome signals around promoter types | FANTOM Consortium 2014. A promoter-level mammalian expression atlas. Nature 507, 462-470, 2014. PMID 24670764 |  |
| Nucleosome organization around YY1 sites | Wang et al. 2012. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors, Genome Research 22, 1798-1812. PMID 22955990 |  |
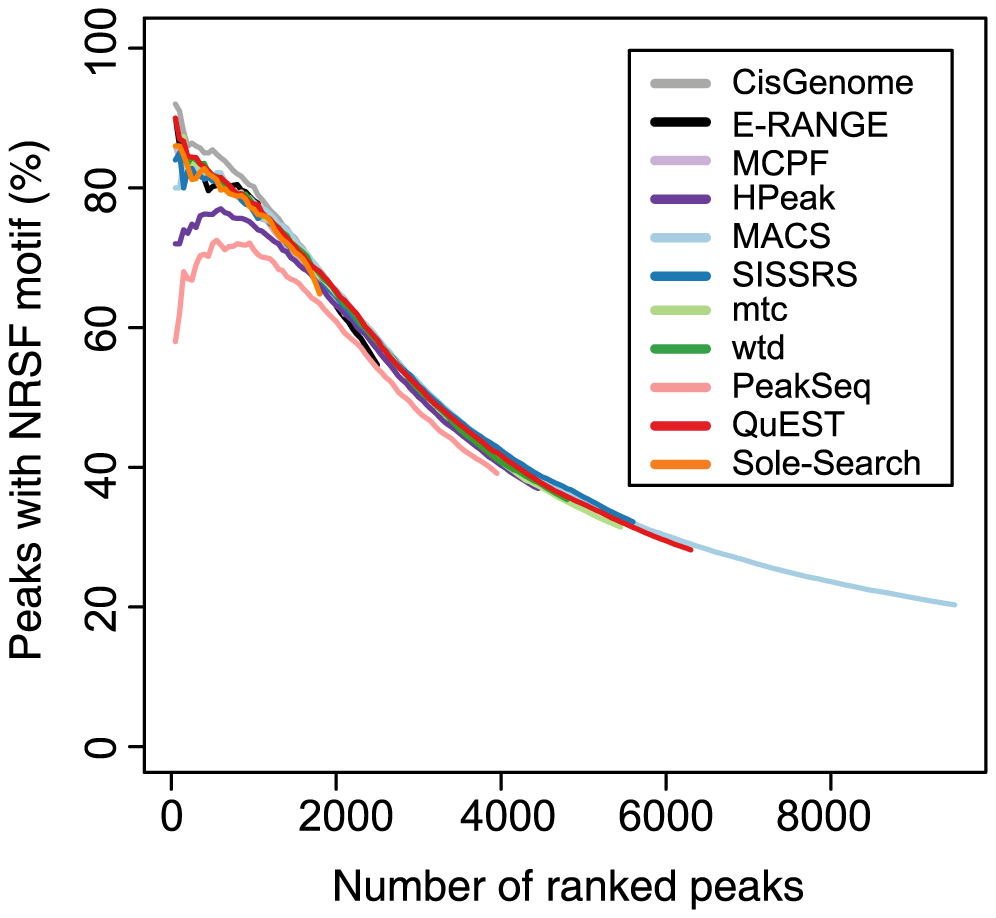
| Peak list evaluation - motif enrichment test | Wilbanks & Facciotti 2010. Evaluation of algorithm performance in ChIP-seq peak detection. PLoS One. 7:e41389. PMID 20628599 |  |