A set of fixed-length sequences located at the same relative position
with respect to a functional site (eg. a transcription start site)
is scanned for occurrences of a given motif. The motif may be defined
by a consensus sequence or a weight matrix. The search may be confined
to a sub-region within the sequences supplied. The program returns either
the sequences containing the motif, the sequences not containing the
motif, or the sequence regions around the motif.
See the
here for
further detail.
MEME Motif Format
The motif library provided by SSA have been originally downloaded from The MEME Suite website. Motifs have then undergone a reformatting process (for more details, please read here).
Matrices from MEME are provided in two formats:
- as letter-probability matrices;
- as integer log-odds weight matrices.
The conversion of base counts into weights is given by the formula shown here:
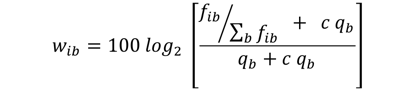 (1)
(1)
where fib is the relative frequency of base b at PWM position i, qb is the background frequency of base b, and c is the fraction of pseudo-counts added to the observed base frequencies.
Unless specified otherwise, background letter frequencies are those from a uniform background (A 0.25000 C 0.25000 G 0.25000 T 0.25000).
Weights are rounded to nearest integers to allow for efficient computation of the probability distribution for scores expected from random sequences.
For more details, visit the MEME website or our PWMLib site .